K-Means LSH
Many of LSH families are structured quantizers: they don’t take into account underlying statistics
- for example, E2LSH is structured:
- we choose only quantization step $w$ and offset $b$ and have little influence on the density of individual cells
- but can address this issue by learning a Vector Quantizer - such as K-Means: this way we can adapt the cell size to the density of the space in the cell
Learning Density with K-Means
Unstructured VQ:
- let $\mathcal R \to [ \, 1, 2, \ … \ , k \, ]$
- and $\mathbf x \to g(\mathbf x) = \mathop{arg min}\limits_{i = 1..k} L_2(\mathbf x, \boldsymbol \mu_i)$
- it maps each vector to a cell indexed by $g(\mathbf x)$
- $k$ is # of possible values of $g(\cdot)$
- $\boldsymbol \mu_i$ are centroids - they define the quantizer
- often learned with K-Means
Illustration:
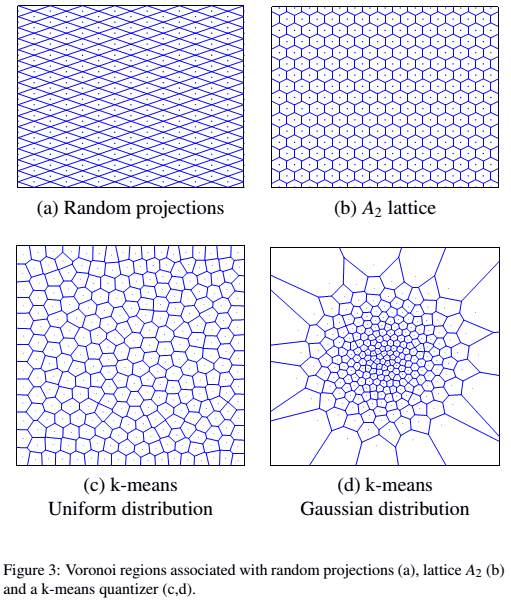
- Euclidean LSH (Random Projection LSH) vs K-Means
- K-Means adapts to data while others don’t
- source: Paulevé2010 figure 3
KLSH: K-Means LSH
Preprocessing
How to use K-Means to build a LSH?
- generate $L$ different clusterings on the same data by using different seeds
- after this we have $L$ codebooks (sets of centroids) ${\mathbf c_{j1}, \ … \ , \mathbf c_{jk}}$, where $\mathbf c_{ji}$ is $i$th centroid of $j$’s clustering
- each centroid is an $h$ and codebooks is an $g$ in LSH
Indexing:
- a vector to index is assigned to the closest centroid found in a codebook
- use $L$ codebooks to have $L$ cluster assignments
Querying
Search time:
- first nearest centroid for each codebook
- then keep only vectors that are assignment to the same centroids
query($\mathbf q$)
- $\text{res} = \varnothing$
- for $j = 1$ to $L$ do
-
$i^* = \operatorname{arg\, mix}\limits_{i = 1 .. k} | \mathbf q, \mathbf c_{ji} |$ - $\text{res} = \text{res} \cup \big{ \text{cluster}(\mathbf c_{ji^*}) \big}$
-
- return $\text{res}$
Improvements
Multi-Probing
Multi-Probing for K-Means LSH:
- fix $m_p$ the number of buckets we want to retrieve
- for each $L$ hash functions
- select $m_p$ closets centroids
- then return all vectors from these $m_p$ centroids
Query-Adaptive K-Means LSH
Idea:
- a variation of Query-Adaptive LSH for K-Means LSH
- instead of a single k-means per hash maintain a pool of independent clustering results
- at the query time select the best one from the pool
Usual Query-Adaptive LSH:
- define a pool of $L$ hash functions (with $L$ larger than in usual LSHs)
- compute relevance criteria $\lambda_j$ for each $g_j$: this criteria identifies the hash functions that are more likely to return the NNs
- relevance could be: distance between the query and the center of the cell
Query-Adaptive K-Means LSH:
-
$\lambda_j(\mathbf q) = \min_{i = 1..k} | \mathbf q, \mathbf c_{ji} |$ - and $\lambda_j(\mathbf q)$ is actually a by-product of finding the nearest centroid - so rank $g_j(\mathbf q)$ by $\lambda_j(\mathbf q)$ then pick $p$ best and use them
Notes:
- for this to be useful need $L$ larger than usual
- it gives better performance, but it becomes more computationally expensive - may be not very critical as it’s done offline
Speeding K-Means Up
Can use Approximate K-Means or Mini-Batch K-Means
Sources
- Paulevé, Loïc, et al. “Locality sensitive hashing: A comparison of hash function types and querying mechanisms.” 2010. [https://hal.inria.fr/inria-00567191/document]